UC Berkeley Press Release
Protein 'yoga' reveals secrets of complex enzyme folding
BERKELEY – University of California, Berkeley, researchers are putting proteins through stretching exercises - what they call "protein yoga" - to uncover the secrets of how proteins fold into elaborate three-dimensional shapes, not unlike the asanas, or postures, of hatha yoga.
A problem that has bedeviled biologists for decades is why a chain of amino acids, akin to a unique sequence of multicolored beads on a string, always folds in a specific way. What are the steps leading to the final, complex molecule, and how does the amino acid sequence determine the ultimate 3-D structure?
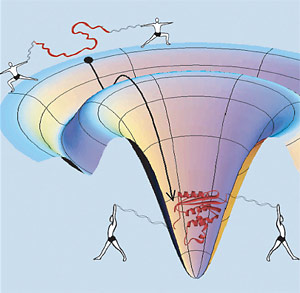 In this whimsical depiction of protein yoga, human figures stand in for the optical tweezers that grab ribbon-like strands of DNA and pull a compact, folded protein (red, center) into its unfolded state (red, along rim). The funnel-like surface depicts the energy landscape traversed by the protein as it relaxes to its native state, which is lowest in energy. Marqusee, Bustamante and colleagues showed that the protein goes through an obligatory partially-folded intermediate (shallow depression) with unusual mechanical properties. (Credit: Elizabeth Shank/UC Berkeley) |
Understanding how proteins fold could lead to the design of new proteins for highly specific functions, or new therapeutic drugs.
UC Berkeley biochemist Susan Marqusee, professor of molecular and cell biology, and biophysicist Carlos Bustamante, professor of physics, chemistry and of molecular and cell biology, decided to investigate this problem one protein at a time. Grabbing both ends of a single protein, they pulled and slowly unbent the protein, then gradually let it relax into its original 3-D shape.
As they put the protein through stretches and relaxations reminiscent of yoga exercises, they verified an experimental observation that has been debated for the past 15 years: In the process of folding, some proteins go through an intermediate state totally unlike their final configuration - a resting state before the final folding occurs.
"People knew that you could have partly-folded proteins, but they're often thought of as being bad - the protein has folded the wrong way and has to come back," Marqusee said. "Such intermediates are also thought to be the precursors for amyloid, the kind of abnormal formations in the brain seen in mad cow disease and Alzheimer's disease.
"Because for the first time we could watch the protein unfold and refold, we could actually see it fold part way and then go from there to the folded protein. It didn't have to step back and try again."
Marqusee, Bustamante and their colleagues - all members of the California Institute for Quantitative Biomedical Research (QB3) at UC Berkeley - reported their findings in the Sept. 23 issue of Science.
Marqusee has studied this particular protein, a bacterial enzyme, in the traditional ways - in test tubes with nuclear magnetic resonance and other forms of spectroscopy - for the past 12 years. Called ribonuclease H, it cleans up little bits of RNA left after the DNA-to-RNA conversions during replication of DNA. Only 155 amino acids long, it is similar to a part of the reverse transcriptase enzyme in the human immunodeficiency virus (HIV) that is a major target of anti-AIDS drugs.
Bustamante, whose lab sits next door, has developed a technique for pulling and twisting single molecules, mostly DNA and RNA, using tools such as optical tweezers to grab the ends of these tiny structures. Eight years ago, he had also grabbed and tugged on a gigantic protein, the muscle protein titin, but it's complexity precluded learning much about its structure and folding.
Marqusee and Bustamante decided to try this technique on RNase H, a much smaller and simpler protein. Because optical tweezers need a large, micron-sized plastic bead to grab hold of, their team decided to use an idea of Bustamante's, that is, to attach molecules of DNA to the protein molecule and use them as points of attachment for the beads. Bustamante's team had successfully used this technique to manipulate small RNAs.
Attaching the protein to a bead "would be like trying to manipulate a red blood cell between two soccer balls, so it made more sense to attach handles to each end of the protein and then grab the ends of these handles," Bustamante said. "Our students Ciro Cecconi and Beth Shank spent more than two years getting the proteins to attach to DNA handles, but finally we got the chemistry right, so it was possible to manipulate a single tiny globular domain of a protein."
During the slow unfolding and refolding, the protein clearly passed through a state unlike the final protein shape, or even a part of that shape. Marqusee referred to this intermediate state as a "molten globule" intermediate because it wasn't rigid like typical folded proteins, but pliant and stretchy. Apparently, this intermediate state makes it easier and faster for the protein to reach its final folded state, and may be necessary for reaching that final, highly precise, 3-D configuration.
"Proteins are like a puzzle, where all the pieces tightly fit together," she said. "They are either completely together, or they are all over the place, unfolded. This intermediate state isn't like most folded proteins, which are brittle; it's compliant, you can pull on it, and it's still in that conformation until you pull on it a lot.
"The intermediate we see in this novel experiment is exactly the same as what we were seeing in the test tube."
Bustamante said that the observations can be explained by distinguishing between secondary interactions among the protein's amino acids, which create structures like helices and sheets, and the tertiary interactions that tie these together into a rigid structure. The molten globule intermediate consists of secondary structures like alpha-helices and beta-sheets, but these aren't yet welded together.
"The molten globule intermediate is collapsed, but has not formed its tertiary interactions that make the folded state - the native state - unique as a very tight, hard, brittle structure," he said. "I think it is very exciting that for the first time we have touched a molten globule. People have talked about molten globules; we have grabbed one and pulled it"
Marqusee, who is the associate director of QB3, sees her collaboration with Bustamante as the epitome of what the institute encourages, and an example of the type of research destined for the new Stanley Hall building nearing completion on the east end of the UC Berkeley campus. The building will be the UC Berkeley headquarters of QB3, which is based at three UC campuses: San Francisco, Santa Cruz and Berkeley.
"This research takes the expertise of two people and, instead of doing something in between, produces something that's really important for both fields," she said.
Marqusee and Bustamante hope to continue their optical tweezer studies of proteins, perhaps with proteins known to fold improperly and aggregate into plaques, such as the amyloid plaques seen in Alzheimer's disease and other neurodegenerative diseases.
The work was supported by grants from the National Institutes of Health, the Department of Energy and the National Science Foundation.

