Gene transcribing machine takes halting, backsliding trip along the DNA
| 30 July 2009
BERKELEY — The body's nanomachines that read our genes don't run as smoothly as previously thought, according to a new study by University of California, Berkeley, scientists.
When these nanoscale protein machines encounter obstacles as they move along the DNA, they stall, often for minutes, and even backtrack as they transcribe DNA that is tightly wound to fit inside the cell's nucleus.
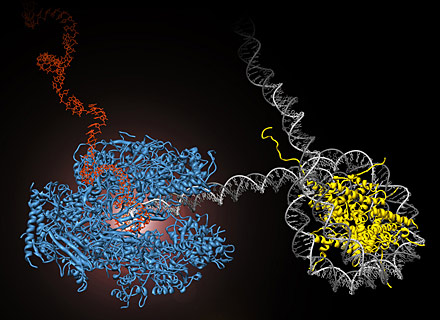 RNA polymerase II (blue) performs the first step of gene expression by moving along the cell’s DNA (gray) and transcribing it into messenger RNA (red). During this process, the polymerase encounters obstacles, such as nucleosomes, which tightly wrap the DNA around histone proteins (yellow) and prevent continued transcription. UC Berkeley researchers have developed methods to directly observe this process in real time. (Courtney Hodges/UC Berkeley)
RNA polymerase II (blue) performs the first step of gene expression by moving along the cell’s DNA (gray) and transcribing it into messenger RNA (red). During this process, the polymerase encounters obstacles, such as nucleosomes, which tightly wrap the DNA around histone proteins (yellow) and prevent continued transcription. UC Berkeley researchers have developed methods to directly observe this process in real time. (Courtney Hodges/UC Berkeley)The measurements, which employed optical tweezers to grab both the polymerase and the end of a single molecule of DNA, are reported in the July 31 issue of the journal Science.
In collaboration with the laboratory of Mikhail Kashlev at the National Cancer Institute, UC Berkeley graduate students Courtney Hodges, Lacra Bintu and their advisor, UC Berkeley's Carlos Bustamante, developed an optical tweezers assay to directly watch individual Pol II complexes as they move along single molecules of DNA. Optical tweezers use laser light to trap and follow a single polymerase in real time, revealing that it truly acts like a biological nanoscale machine as it moves along our genes.
The main obstacle to smooth operation of Pol II is the nucleosome, a bundle of eight histone proteins around which DNA wraps tightly. Tens of thousands of nucleosomes are bundled together into a chromosome, efficiently packaging six feet of DNA into a nucleus a million times smaller. The researchers were able to place a single nucleosome in front of the polymerase and then use the optical tweezers to observe what happens when the polymerase encounters this roadblock.
"For over 30 years, scientists had wondered how the polymerase responded to the nucleosome, and we were finally able to observe this process directly," Hodges said. "People thought that the polymerase is a powerful motor that would blow through the nucleosome like a bulldozer, but it's surprisingly delicate in its response; if anything is in the way, Pol II stops and backs up."
Bintu noted that this halting movement – 20-50 steps forward, then a couple of steps back – could be a key part of how gene expression is regulated. Nucleosomes are highly regulated by other proteins and can provide signals that control Pol II, much like a traffic light regulates street traffic, she said. Regulatory proteins may bind to the nucleosome and make the DNA unwind more easily, or could latch onto Pol II and prevent it from backsliding. Either would speed up transcription, while regulatory proteins that compact DNA and nucleosomes further slow down or even stop transcription.
Scientists have for years imagined that nucleosomes must be "loosened up" to allow for gene expression, and the authors note that their results give a more detailed, mechanistic insight into this process.
"Our study indicates that modulation of the wrapping/unwrapping equilibrium of DNA around the histone octamer constitutes the physical basis for regulation of transcription through nucleosomal DNA," the authors wrote.
On the flip side, disturbances in nucleosome regulation could lead to disease.
"Transcription is a central point of control for gene expression, since everything from coordination of development to prevention of uncontrolled cell growth, that is, cancer, involves a highly regulated program of transcription by Pol II," Bintu said. "When transcription goes haywire, pathologies like cancer and developmental abnormalities usually follow."
Hodges and Bintu compare the DNA in the nucleosome to a band of sticky Velcro looped a couple of times around the histone proteins. The DNA is constantly being pushed around, however, and tends to peel off and then reattach to the histones. When the DNA is bound to the histones, Pol II cannot read it and transcription pauses. The polymerase restarts transcription only when the DNA briefly comes off the histones and, acting like a ratchet, works its way along the DNA throughout the entire nucleosome. At some point, the nucleosome leapfrogs over Pol II and the nanomachine trundles along unhindered.
The researchers also tugged on the two ends of a DNA molecule after transcription to see what had happened to the nucleosome. They found that the nucleosome was frequently ejected from the DNA because the tension prevented the DNA from forming loops that would have allowed the nucleosome to skip over Pol II.
"We found that even a very small amount of tension in the DNA – 3 to 5 piconewtons – during transcription results in Pol II removing the nucleosome from DNA like a pair of wire strippers," Hodges said. "It's very likely that the DNA in our bodies is very taut at some places and loose in others, so we think it's possible that the cell uses tension in the genome to alter the dynamics of nucleosomes in certain genes."
"These experiments give a much more dynamic picture of the nucleosome, showing that it isn't a static bead-on-a-string but an active structure that can regulate when and how our genetic information is read," Bintu said. "This is just one single nucleosome, but it is the first step in understanding epigenetic effects that make one cell behave differently from another."
Hodges is in the biophysics graduate group, and Bintu is a graduate student in physics. Both are part of Bustamante's Jason L. Choy Laboratory of Single-Molecule Biophysics, named after a chemistry graduate student who died in an automobile accident in 2005. Bustamante is a professor of physics, chemistry and of molecular and cell biology, a Howard Hughes Medical Institute investigator and an affiliate of the California Institute for Quantitative Biosciences (QB3).
The work is supported by the National Institutes of Health.

