Chromosomes dance and pair up on the nuclear membrane
| 12 November 2009
BERKELEY — Meiosis – the pairing and recombination of chromosomes, followed by segregation of half to each egg or sperm cell – is a major crossroads in all organisms reproducing sexually. Yet, how the cell precisely choreographs these chromosomal interactions is a long-standing question.
In fact, the cytoskeleton appears to encourage the dance of the chromosomes around the nuclear membrane as they search for their partners, and help make sure they have the right partner before meiosis continues.
The results are published online today (Nov. 12), and will appear in the Nov. 25 print issue of the journal Cell.
The new finding in the nematode worm C. elegans match related but less detailed findings in organisms ranging from wheat and fungi (yeast) to flies and mice, suggesting that the basic mechanics of sex – at least at the cellular level – arose more than a billion years ago when single- celled organisms first leaned how to mix and match their genes as a strategy for survival.
"Errors during meiosis lead to age-related human infertility, and to birth defects such as Down syndrome and Klinefelter syndrome," said Abby Dernburg, UC Berkeley associate professor of molecular and cell biology and a Howard Hughes Medical Institute investigator. "Our work teaches us about the fundamental mechanisms of genome organization, about how cells execute processes in precise ways, monitor their own mistakes and correct or eliminate them."
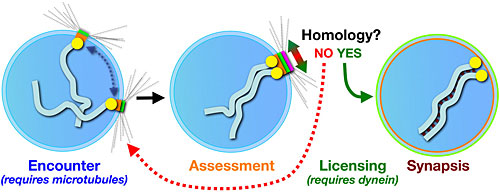 This simple model of a nucleus with only one pair of chromosomes illustrates the process of synapsis – the pairing of homologous chromosomes. In early meiosis, the chromosomes attach by their pairing centers to proteins on the nuclear envelope, which are linked to the cytoskeleton of the cell. The microtubules in the cytoskeleton facilitate movement of the patches and associated chromosomes, promoting encounters between chromosomes. Once the chromosomes come together, a protein called dynein assesses whether or not the chromosomes are homologous and, if yes, allows formation of a zipper-like synaptonemal complex between the two.
This simple model of a nucleus with only one pair of chromosomes illustrates the process of synapsis – the pairing of homologous chromosomes. In early meiosis, the chromosomes attach by their pairing centers to proteins on the nuclear envelope, which are linked to the cytoskeleton of the cell. The microtubules in the cytoskeleton facilitate movement of the patches and associated chromosomes, promoting encounters between chromosomes. Once the chromosomes come together, a protein called dynein assesses whether or not the chromosomes are homologous and, if yes, allows formation of a zipper-like synaptonemal complex between the two.Dernburg studies the first steps in meiosis; steps that, in humans, are completed in the ovaries of a female well before she is born. This involves the pairing, or synapsis, of homologous chromosomes in which the chromosome from the mother pairs with the analogous chromosome from the father. In humans, that means 46 chromosomes pair off into 23, but in the nematode, 12 chromosomes pair off into 6.
To stabilize the chromosome pairs, protein links form along the length of the homologs, like a zipper. Called the synaptonemal complex, this zipper seems necessary to allow the homologues to break and recombine, thereby exchanging a set of genes between Mom and Dad before sending the chromosomes into the world aboard egg or sperm.
"The whole goal of the cell at this developmental stage is to pair up homologous chromosomes, to reinforce that pairing through formation of the synaptonemal complex, to make crossovers between homologs, and then to separate the pairs into different daughter cells," said Dernburg, who is also a researcher at Lawrence Berkeley National Laboratory (LBNL) and a faculty affiliate of the California Institute for Quantitative Biosciences (QB3). "But the synaptonemal complex can form between non-homologous regions of the chromosomes, so pairing has to be coordinated with synapsis, and synapsis has to be regulated so it happens only between homologous chromosomes."
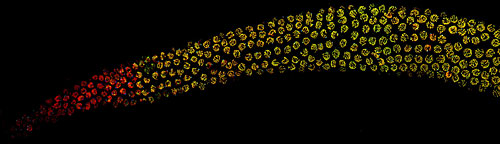 The gonads of a nematode make up about half of an adult organism's cells, and contain both sperm and eggs at various stages of development in a temporal progression. The cells with only red staining have not yet entered meiosis, while the cells stained both red and green have begun or completed chromosome pairing and synapsis. Each worm completes its life cycle in just three days, producing up to 200 offspring.
The gonads of a nematode make up about half of an adult organism's cells, and contain both sperm and eggs at various stages of development in a temporal progression. The cells with only red staining have not yet entered meiosis, while the cells stained both red and green have begun or completed chromosome pairing and synapsis. Each worm completes its life cycle in just three days, producing up to 200 offspring.To many biologists, it seemed likely that synaptonemal complex formation would occur spontaneously between paired chromosomes, Dernburg said. But she and her colleagues have now shown that the cell actively prevents formation of the synaptonemal complex until it has checked and made sure the paired chromosomes are really homologs.
"The punch line of our paper is that it indicates there is a check point that we weren't aware of, and which we believe is testing homology between paired chromosomes and licensing the formation of the synaptonemal complex when it is satisfied that the chromosomes are homologous pairs," she said. "It probably facilitates the separation of mispaired chromosomes and helps them find their right partner."
She also showed that a motor protein called dynein is necessary for this checkpoint mechanism.
"When you get rid of dynein, the homologs pair, though more slowly, but they don't go on to form the synaptonemal complex, suggesting that dynein overcomes an inhibitory mechanism," she said. "Mutations in nuclear envelope components that link to the cytoskeleton show that the barrier to synapsis is imposed by the connection between the chromosomes and these nuclear envelope proteins."
There are indications that the constraints on chromosome pairing involve mechanical stresses imposed by the connection between the cytoskeleton, the nuclear envelope proteins and the chromosomes, and that the pull from the nuclear envelope releases a constraint that tells the chromosomes that it is OK to synapse.
Despite obvious differences between nematodes and humans, Dernburg noted, the mechanism of synapsis control is probably very ancient.
"It is probably highly conserved that chromosomes attach to the nuclear envelope during meiosis, though they usually do it through their telomeres," she said. "At the nuclear envelope, the chromosomes attach to a force-generating mechanism, and C. elegans has probably just evolved a variant of this."
Dernburg continues to explore the protein complexes in the nuclear membrane that coordinate synapsis, and the protein zipper that connects homologs during synapsis.
Dernburg's coauthors are UC Berkeley graduate students Aya Sato, Regina Rillo, David Wynne and Roshni A. Kasad; Berith Isaac, formerly of LBNL and now at Harvard University; Carolyn M. Phillips, who recently obtained her UC Berkeley Ph.D. and moved to Massachusetts General Hospital; and Peter M. Carlton, from the laboratory of John Sedat at UC San Francisco.
The work was supported by the Howard Hughes Medical Institute, and by research grants from the American Cancer Society and the National Institute of General Medical Sciences of the National Institutes of Health.

